Research Projects
Our research focuses upon developing cutting-edge computational technologies for use in patient-specific cancer simulators. Our ultimate goal is to create quantitative platforms that integrate patient data from multiple sources (proteomics, histopathology, radiology), to help guide surgical and therapeutic planning.
Core Technologies

PhysiCell: Physics-based cell model
This core technology, published in PLoS Computational Biology in 2018 (and building a decade of prior development), models individual cells as agents. Each agent has a lattice-free position (center of mass), a velocity that is determined by the balance of biomechanical forces, and a phenotype that depends upon the cell's internal genomic/proteomic state and its sampling of the local microenvironment. Cell cycle progression and apoptotic and necrotic cell death are modeled as stochastic processes that vary with the microenvironment. Detailed, state-dependent "submodels" regulate the cell's fluid and biomass content. We are the first to model the critical process of nuclear cell calcification, and have the most detailed model of cell necrosis.
This agent model is the foundation of our cutting-edge patient calibration techniques, which can be uniquely constrained to patient immunohistochemistry and other histopathologic (e.g., morphometric) measurements. PhysiCell is capable of simulating 105 to 106 cells on desktop processors in complex 3-D tissue structures.
More information: PhysiCell.org
Recent reference: Ghaffarizadeh et al. (2018)
Recent multimedia: immune cells attacking a 3-D tumor,
bioengineered cell-based cancer therapy,
DCIS simulations
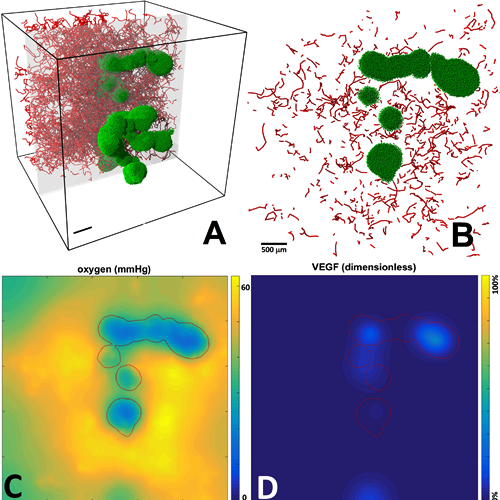
BioFVM: A biological transport solver
This code simulates the diffusive transport of multiple substrates
(e.g., oxygen, glucose, growth factors, drugs) as they are secreted/released/absorbed
by biological elements in 3-D tissues. We use this code to simulate the microenvironment
and its interactions with cells. BioFVM is capable of simulating diffusive transport of
5-10 substrates on 1-10 million voxel
meshes (generally 5-100 mm3) on desktop processors.
Click here to read more.
BioFVM has been peer reviewed in
Bioinformatics.
It is
available as open source under the (3-clause) BSD license.

Recent reference: Ghaffarizadeh et al. (2016)
Project website: BioFVM.MathCancer.org
Alternate download site: click here
Tissue-scale model
Our continuum tissue-scale model simulates the tumor-host interface as a moving boundary problem, using advanced level set techniques that can automatically handle changes in the tumor shape (e.g., splitting into fragments, developing invasive fingers that can merge or split, etc.). Substrate transport is coupled using nonlinear reaction-diffusion equations. We have successfully tied this simulation to models of angiogenesis and blood flow.
The simulator is capable of modeling tumor growth in complex, heterogeneous 2-D tissues at large spatial scales (approximately 1 cm) on a single CPU. This model was originally developed from 2001 to 2007, and has been superceded (for the time being) by agent-based modeling.
Recent references:
Macklin and Lowengrub (2007),
Frieboes et al. (2007),
Macklin and Lowengrub (2008),
Macklin et al. (2009)
Recent multimedia: Coupled tumor-angiogenesis simulation
intracranial tumor simulations

MultiCellDS
MultiCellDS (Multicellular data standard), an outgrowth of the earlier MultiCellXML project, aims to create a data standard for sharing multicellular experimental, simulation, and clinical data. Our ultimate goal is to foster a community that develops user-friendly tools that can read, write, and recombine data into better simulations and analyses for multicellular biology and predictive medicine. As part of this effort, we are developing MultiCellDB: a repository for a curated libary of digital cell lines and peer-reviewed simulation and experimental data.
A novel part of MultiCellDS is the digital cell line: a digital analogue of experimental cell lines that will help to collect biophysical cell line measurements coming from many research groups and make them readily accessible to an ecosystem of compatible computational models. Digital snapshots provide a unified, model-independent representation of simulation data, as well as segmented pathology, radiology, and experimental data.
In the past year, MultiCellDS has grown from a single-lab effort to an international community of mathematicians, biologists, data scientists, and clinicians, with contributors in the US, the United Kingdom, Germany, and elsewhere. We are currently preparing our first method paper on a repository of over 200 digital cell lines, many of which were created from brain and breast cancer patient data. Click here to read more!
Project website: MultiCelDS.org

